Which Best Describes the Antisense Technique
The human genome is a complete set of nucleic acid sequences for humans encoded as DNA within the 23 chromosome pairs in cell nuclei and in a small DNA molecule found within individual mitochondriaThese are usually treated separately as the nuclear genome and the mitochondrial genome. Ule and colleagues discuss cross-linking and immunoprecipitation CLIP methods for characterizing the RNA binding partners of RNA-binding proteins and explore the data analysis workflows best.

Antisense Technology An Overview Sciencedirect Topics
During the primer extension primers 2a3a1s bound.
. An applicant need not have actually reduced the invention to practice prior to filing. 6 to 30 characters long. They are used to detect and destroy DNA from similar.
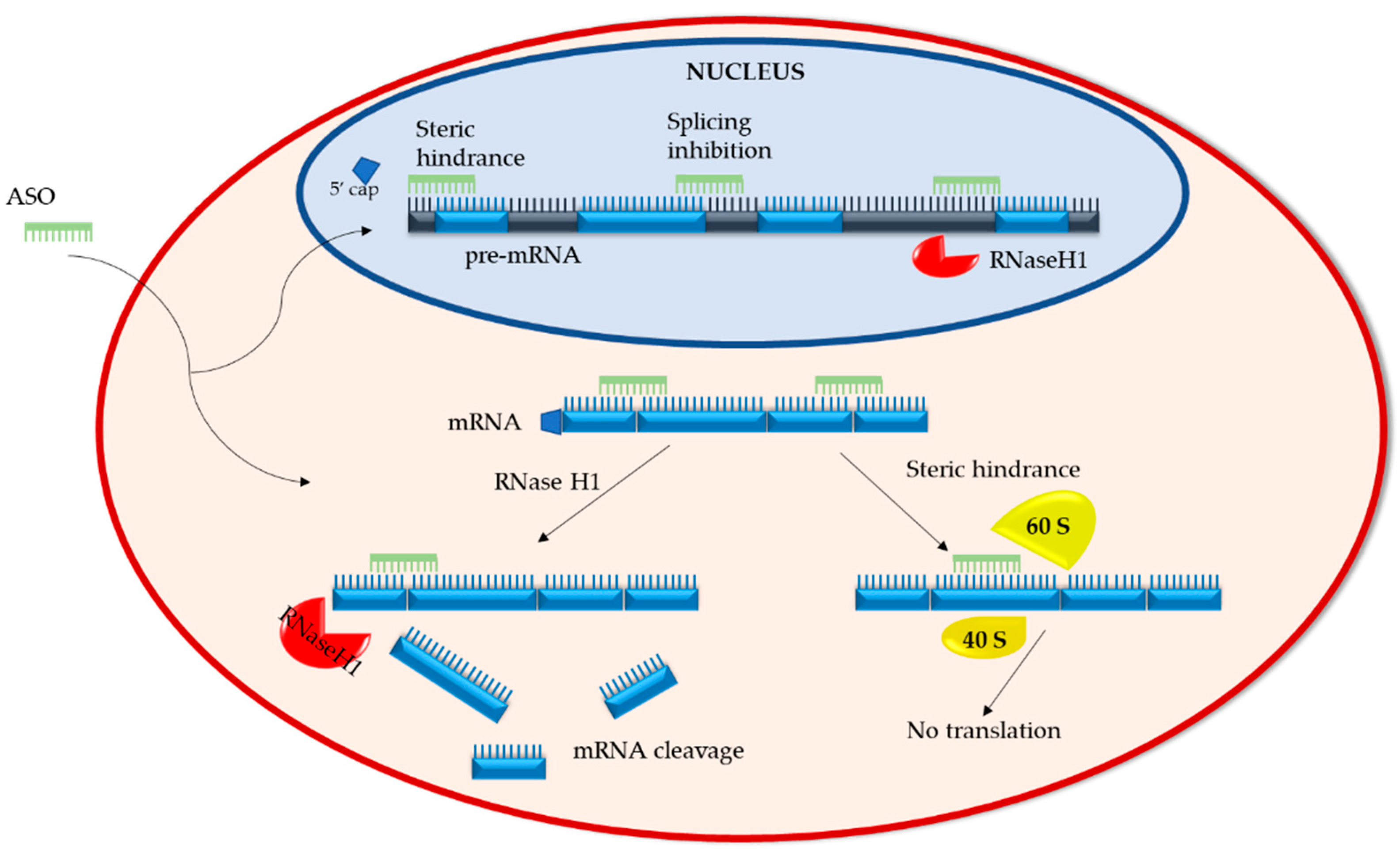
This technique does not require any nucleic acid purification or amplification steps to detect its target RNA rather it involves unique antisense oligonucleotides that can bind to SARS-CoV-2 RNA with similar efficiency irrespective of its current mutations. In Arabidopsis and other plants directed gene silencing has often been approached by RNAi. Quigg 822 F2d 1074 1078 3 USPQ 2d 1302 1304 Fed.
2003 for natural miRNA targets. We showed that the same two N gene targeted ASOs can successfully be used for a point of care POC rapid cost-effective and. Whereas initial experiments using overexpression of antisense transcripts antisense to the gene which should be silenced had only limited.
The antisense strand product can therefore be extended after self-folding from the 3a site by adding a 4a or 4a5a sequence at the loop region. A prophetic example describes an embodiment of the invention based on predicted results rather than work actually conducted or results actually achieved. Hairpin RNAi versus artificial microRNA.
1987 as of Goulds filing date no person had built a. ASCII characters only characters found on a standard US keyboard. This technique has been first described by Palatnik et al.
CRISPR ˈ k r ɪ s p ər an acronym for clustered regularly interspaced short palindromic repeats is a family of DNA sequences found in the genomes of prokaryotic organisms such as bacteria and archaea. Must contain at least 4 different symbols. Human genomes include both protein-coding DNA genes and noncoding DNA.
These sequences are derived from DNA fragments of bacteriophages that had previously infected the prokaryote.

Biomedicines Free Full Text The Challenges And Strategies Of Antisense Oligonucleotide Drug Delivery Html

Antisense Transcription Dependent Chromatin Signature Modulates Sense Transcription And Transcript Dynamics Biorxiv

The Powerful World Of Antisense Oligonucleotides From Bench To Bedside Quemener 2020 Wires Rna Wiley Online Library

Rna Centric Methods Toward The Interactome Of Specific Rna Transcripts Trends In Biotechnology
No comments for "Which Best Describes the Antisense Technique"
Post a Comment